Pipeline Configuration¶
Create the SQLite database: The openVA Pipeline uses an SQLite database, called the Transfer DB, to store and access configuration settings for ODK Central, openVA, and DHIS2. Error and log messages are also stored to this database, along with the VA records downloaded from ODK Central and the assigned COD.
While it is possible to create the Transfer DB manually (see the next bullet point) the openva-pipeline package has a built-in function for creating the database with the default settings. Open a terminal shell, change to the Pipeline’s working directory, and start a Python session in the virtual environment with the following commands:
$ source venv/bin/activate $ python
Within the Python interpreter load the openVA Pipeline package and call the
create_transfer_db():>>> import openva_pipeline as ovapl >>> ovapl.create_transfer_db('Pipeline.db', '.', 'enilepiP') >>> quit()
This will create an encrypted SQLite database, called Pipeline.db, in the working directory with encryption key enilepiP. (Exit the virtual environment with
deactivate)Manual Installation
The necessary tables and schema are created in the SQL file pipelineDB.sql, which can be downloaded from the OpenVA_Pipeline GitHub webpage. Create the SQLite database in the folder that will serve as the Pipeline’s working directory.
Use SQLCipher to create the Pipeline database, assign an encryption key, and populate the database using the following commands (note that the
$is the terminal prompt andsqlite>is the SQLite prompt, i.e., not part of the commands).
$ sqlcipher sqlite> .open Pipeline.db sqlite> PRAGMA key="encryption_key"; sqlite> .read "pipelineDB.sql" sqlite> .tables sqlite> -- take a look -- sqlite> .schema ODK_Conf sqlite> SELECT odkURL from ODK_Conf; sqlite> .quit
- Note how the Pipeline database is encrypted, and can be accessed via with SQLite command:
PRAGMA key = "encryption_key;"
$ sqlcipher sqlite> .open Pipeline.db sqlite> .tables Error: file is encrypted or is not a database sqlite> PRAGMA key = "encryption_key"; sqlite> .tables sqlite> -- update tables as follows -- sqlite> INSERT INTO ODK_Conf (odkURL, odkUser) VALUES ("http://your.odk.server.address", "your_odk_user_name"); sqlite> -- look at changes -- sqlite> SELECT odkURL, odkUser from ODK_Conf; sqlite> .quit
Configure Pipeline: The Pipeline connects to ODK Central (or Central) and DHIS2 servers and thus requires usernames, passwords, and URLs. Arguments for openVA should also be supplied. We will use DB Browser for SQLite to configure these settings. Start by launching DB Browser from the terminal, which should open the window below
$ sqlitebrowser
Next, open the database by selecting the menu options: File -> Open Database…
and navigate to the Pipeline.db SQLite database and click the Open button. This will prompt you to enter in encryption password.
- ODK Configuration: To configure the Pipeline connection to ODK Central, click on the Browse Data tab and
select the ODK_Conf table as shown below.
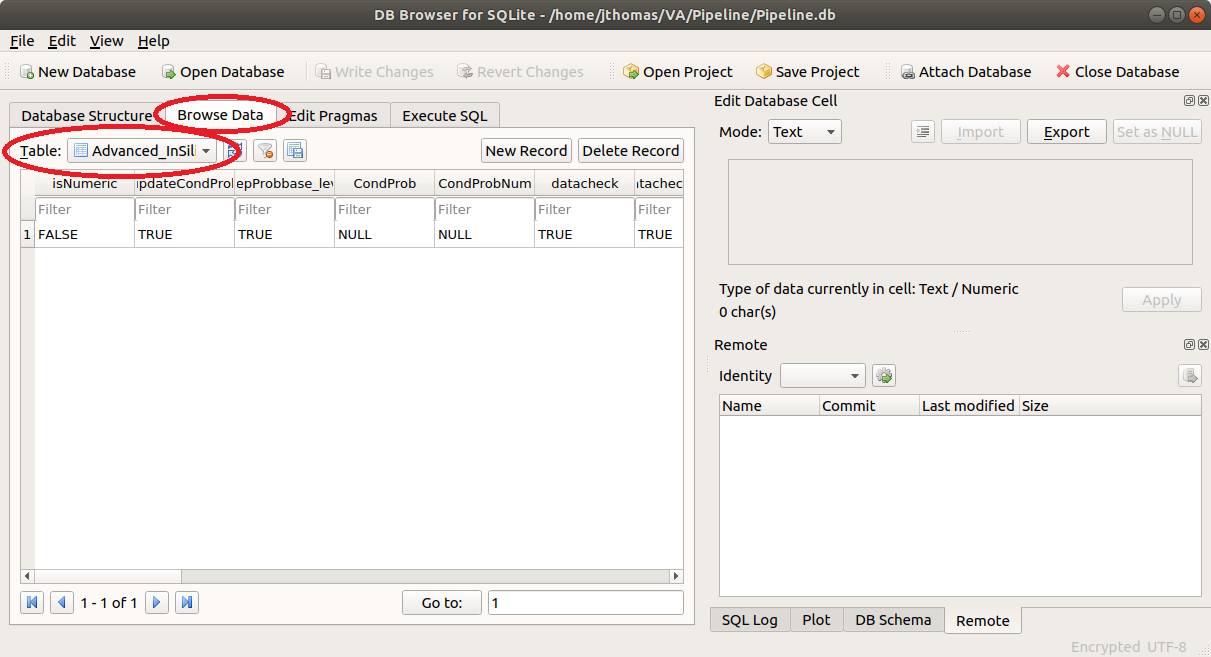
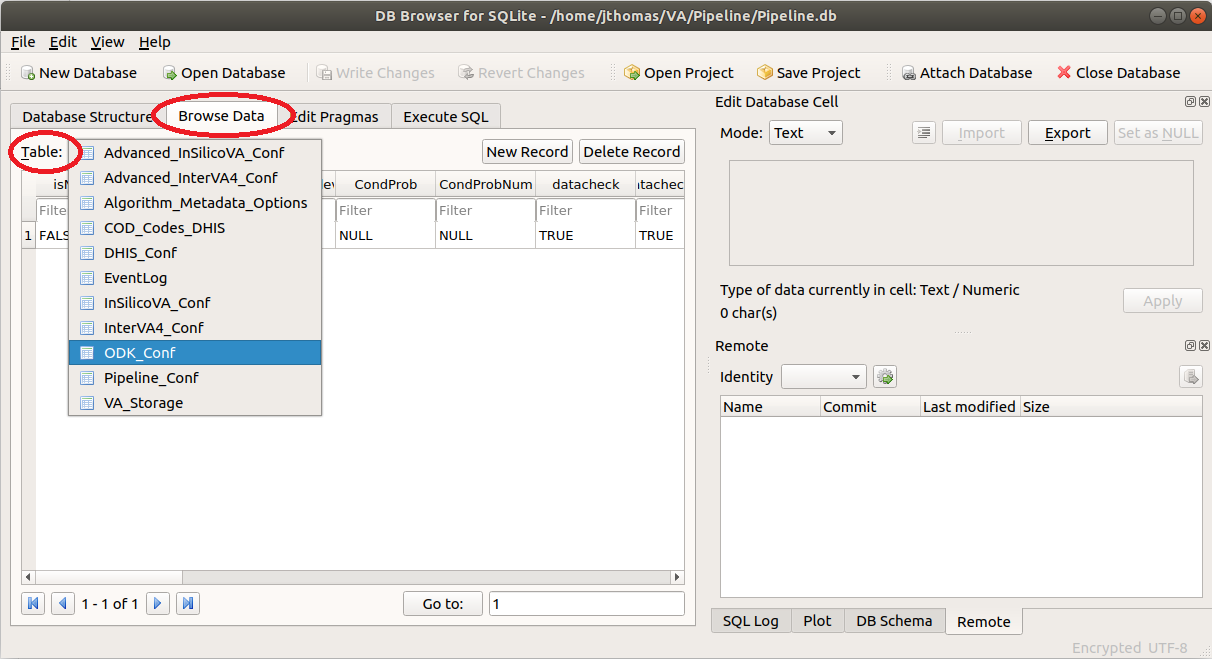
Now, click on the odkURL column, enter the URL for your ODK Central server, and click Apply.
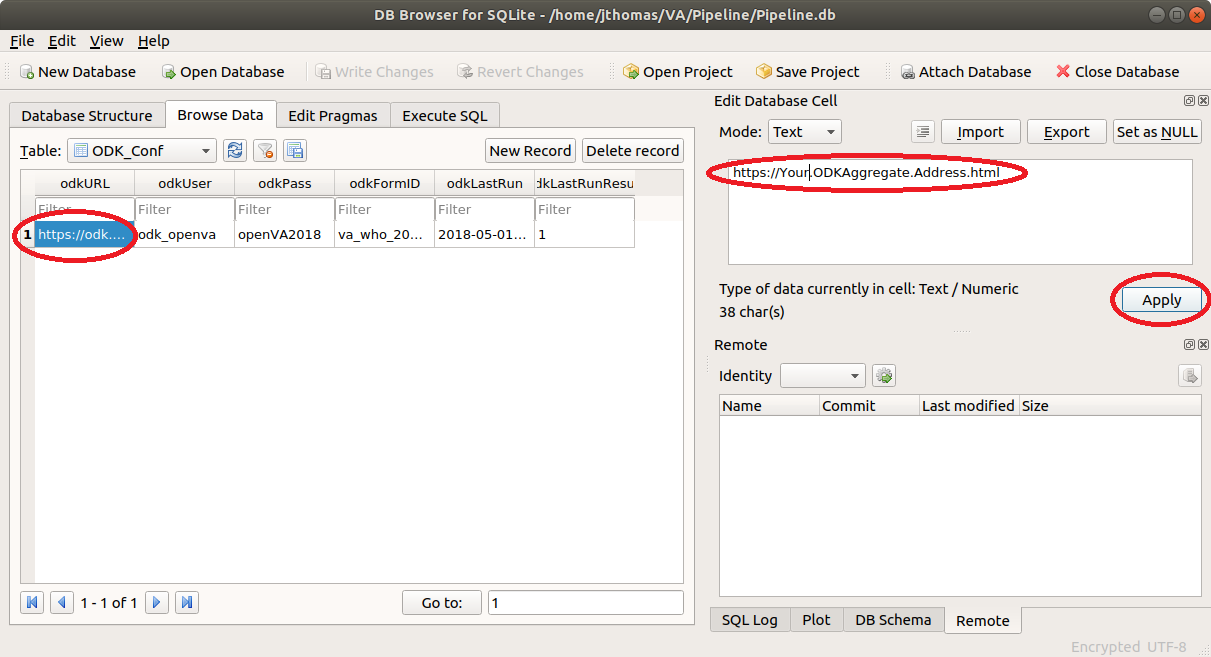
Similarly, edit the odkUser, odkPass, and odkFormID columns so they contain a valid user name, password, and Form ID (see Form Management on ODK Central server) of the VA questionnaire of your ODK Central server.
- Configure ODK_Conf table from a Terminal:
(note that the
$is the terminal prompt andsqlite>is the SQLite prompt, i.e., not part of the commands).
$ sqlcipher sqlite> .open Pipeline.db sqlite> PRAGMA key="encryption_key"; sqlite> .read "pipelineDB.sql" sqlite> .tables sqlite> -- take a look -- sqlite> .schema ODK_Conf sqlite> SELECT odkURL from ODK_Conf; sqlite> .quit
- openVA Configuration: The Pipeline configuration for openVA is stored in the Pipeline_Conf table. Follow
the steps described above (in the ODK Central Configuration section) and edit the following columns:
workingDirectory – the directory where the Pipeline files (i.e., Pipeline.db) are stored. Note that the Pipeline will create new folders and files in this working directory, and must be run by a user with privileges for writing files to this location.
algorithm – currently, there are only three acceptable values for the algorithm:
InSilicoVA,InterVAorSmartVAalgorithmMetadataCode – this column captures the necessary inputs for producing a COD, namely the VA questionnaire, the algorithm, and the symptom-cause information (SCI) (for more details, see the section: Symptom-Cause Information). Note that there are also different versions (e.g., InterVA 4.01 and InterVA 4.02, or WHO 2012 questionnaire and the WHO 2016 instrument/questionnaire). It is important to keep track of these inputs in order to make the COD determination reproducible and to fully understand the assignment of the COD. A list of all algorith metadata codes is provided in the dhisCode column in the Algorithm_Metadata_Options table. The logic for each code is
algorith|algorithm version|SCI|SCI version|instrument|instrument version
codSource – both the InterVA and InSilicoVA algorithms return CODs from a list produced by the WHO, and thus this column should be left at the default value of
WHO.
- DHIS2 Configuration: The Pipeline configuration for DHIS2 is located in the DHIS_Conf table, and the
following columns should be edited with appropriate values for your DHIS2 server.
dhisURL – the URL for your DHIS2 server
dhisUser – the username for the DHIS2 account
dhisPass – the password for the DHIS2 account
dhisOrgUnit – the Organization Unit (e.g., districts) UID to which the verbal autopsies are associated. The organisation unit must be linked to the Verbal Autopsy program. For more details, see the DHIS2 Verbal Autopsy program installation guide Alternatively, if there are columns in your ODK form that identify the organization unit of each VA record, then include the column names in this field (e.g., “Region, District, Tract”). For this option, simply use the final part of the ODK column name. For example, if a column is labeled “consented-deceased_CRVA-in_on_deceased-Region”, only include the last part (Region) in the dhisOrgUnit field.
SmartVA Configuration: The Pipeline can also be configured to run SmartVA using the command line interface (CLI) available from the ihmeuw/SmartVA-Analyze repository.
#. Download the smartva CLI from the following repository: https://github.com/ihmeuw/SmartVA-Analyze/releases and save it in the Pipeline’s working directory (see below).
Update the Pipeline_Conf table in the SQLite database with the following values:
workingDirectory – the directory where the Pipeline files are stored – THIS IS WHERE THE smartva CLI file should be downloaded.
openVA_Algorithm – set this field to
SmartVAalgorithmMetadataCode – set this field to the appropriate SCI, e.g.
SmartVA|2.0.0_a8|PHMRCShort|1|PHMRCShort|1
codSource – set this field to``Tariff``.
Miscellaneous Notes¶
Symptom-Cause Information¶
A key component of automated cause assignment methods for VA is the symptom-cause information (SCI) that describes how VA symptoms are related to each cause. It is likely that the relationships of VA symptoms to causes vary in important ways across space and between administrative jurisdictions, and they are likely to change through time as new diseases and conditions emerge and as treatments become available. Consequently, automated cause assignment algorithms used for mortality surveillance should optimally rely on representative SCI that is locally and continuously updated. Furthermore, it is vital to track the SCI used for COD assignment to enable reproducibility and to fully understand the assignment of the COD.